Our paper was accepted in EHJ Open!
Abstract
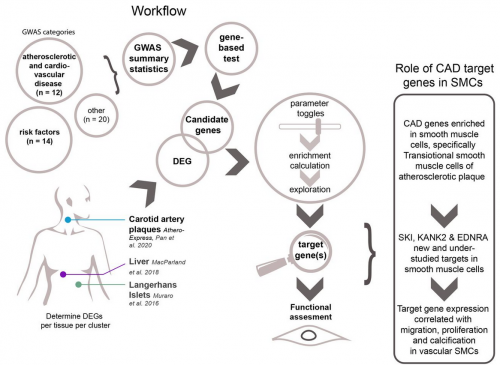
Background Genome-wide association studies have discovered hundreds of common genetic variants for atherosclerotic disease and cardiovascular risk factors. The translation of susceptibility loci into biological mechanisms and targets for drug discovery remains challenging. Intersecting genetic and gene expression data has led to the identification of candidate genes. However, previously studied tissues are often non-diseased and heterogeneous in cell composition, hindering accurate candidate prioritization. Therefore, we analyzed single-cell transcriptomics from atherosclerotic plaques for cell-type-specific expression to identify atherosclerosis-associated candidate gene-cell pairs.
Methods and Results To identify disease-associated genes, we applied gene-based analyses using GWAS summary statistics from 46 atherosclerotic and cardiovascular disease, risk factors, and other traits. We then intersected these candidates with scRNA-seq data to identify genes specific for individual cell (sub)populations in atherosclerotic plaques. The coronary artery disease loci demonstrated a prominent signal in plaque smooth muscle cells (SKI, KANK2, SORT1) p-adj. = 0.0012, and endothelial cells (SLC44A1, ATP2B1) p-adj. = 0.0011. Further sub clustering revealed genes in risk loci for coronary calcification specifically enriched in a synthetic smooth muscle cell population. Finally, we used liver-derived scRNA-seq data and showed hepatocyte-specific enrichment of genes involved in serum lipid levels.
Conclusion We discovered novel gene-cell pairs, on top of known pairs, pointing to new biological mechanisms of atherosclerotic disease. We highlight that loci associated with coronary artery disease reveal prominent association levels in mainly plaque smooth muscle and endothelial cell populations. We present an intuitive single-cell transcriptomics-driven workflow rooted in human large-scale genetic studies to identify putative candidate genes and affected cells associated with cardiovascular traits. Collectively, our workflow allows for the identification of cell-specific targets relevant for atherosclerosis and can be universally applied to other complex genetic diseases and traits.
Translational perspective GWAS identified a large number of genomic loci associated with atherosclerotic disease. The translation of these results into drug development and faster diagnostics remains challenging. With our approach, we cross-reference the GWAS findings for atherosclerotic disease with scRNA-seq data of disease-relevant tissue and bring the GWAS findings closer to the functional and mechanistic studies.
Full text is here.


Great content! Keep up the good work!